Taming the evolution – a short tale on the directed evolution of enzymes
Ewa Kot 1, Katarzyna Kurpiewska 2, Maciej Szaleniec 1
1 Jerzy Haber Institute of Catalysis and Surface Chemistry, Polish Academy of Sciences, Kraków
2 Faculty of Chemistry, Jagiellonian University, Kraków
Abstract
Evolution is an amazing process that, during billions of years, led to the transformation of primitive cells of primordial life into an incredibly complex and biodiverse ecosystem of our planet. In the process of evolution, all the components that build living cells have changed, including protein enzymes that accelerate chemical reactions. Recently, enzymes are more and more readily used in industry. Molecular biology and genetic engineering provide means for enzyme modification that results in the development of new catalytic capabilities with no counterpart in living organisms. One of the more promising methods of such modifications is directed enzyme evolution. In recognition of the groundbreaking importance of molecular evolutionary techniques in science, on 3rd October 2018, the Royal Swedish Academy of Sciences announced the winners of this year’s Nobel Prize in Chemistry. The award for “use of the mechanisms of evolution” was divided into two parts – the first part was awarded to Frances H. Arnold of Caltech, California, the second to George P. Smith from the University of Missouri, Columbia, and Sir Gregor P. Winter from the MRC Laboratory of Molecular Biology in Cambridge, UK. The directed enzyme evolution method developed by Professor Arnold, the fifth woman in history to win a Nobel Prize, is presented in this article.
Introduction
On 3rd October 2018, the Royal Swedish Academy of Sciences announced the winners of this year’s Nobel Prize in Chemistry. The award for “exploiting the mechanisms of evolution” was divided into two parts – the first part was awarded to Frances H. Arnold of Caltech, California, the second to George P. Smith from the University of Missouri, Columbia, and Sir Gregory P. Winter from the MRC Laboratory of Molecular Biology in Cambridge, UK. The first part of the Award concerned the research on the directed evolution of enzymes. The method developed by Professor Arnold, the fifth woman in history to be awarded a Nobel Prize, has made a breakthrough in the industrial application of enzymes over the past two decades. This article is devoted to an introduction to this revolutionary method.
Enzymes are proteins that are responsible for accelerating and controlling most of the chemical processes in living organisms. Currently, they are readily used in industry as biocatalysts accelerating chemical reactions. Very often they exhibit higher efficiency and selectivity in chemical transformations than conventional catalysts. However, only a small part of natural enzymes have features desired by industry i.e, resistance to high temperatures and the presence of organic solvents, or the possibility of using the same portion of the enzyme in several reaction cycles [9]. We also very often want to use enzymes to catalyze the transformation of unusual chemical molecules that have no counterpart in living organisms – unfortunately, natural enzymes do not recognize such molecules! The most direct way to obtain enzymes with the desired properties would be the so-called rational redesign of proteins, i.e. such modification of their amino acid sequence that would allow obtaining the desired features. For the rational redesign, however, it is necessary to know the relationship between the amino acid sequence (stored in the genes carrying information about the enzyme), and its structure and catalytic properties. The knowledge of such relationships is still available only for a small percentage of enzymes that can be useful in the industry [7]. The solution to this problem is “directed evolution” – a technique that allows the modification of enzymes in the desired direction.
From DNA to enzyme
Before we go into explaining the ins and outs of the directed evolution of enzymes, there are some basic concepts in molecular biology that need to be clarified. Enzymes are proteins that are responsible for accelerating and controlling most chemical processes. The central dogma of molecular biology, a theory that has been accepted unconditionally and cannot be challenged, assumes that proteins are the ultimate physical form of information contained in DNA (deoxyribonucleic acid) (Fig. 1). Roughly, it could be compared to baking a cake. The cookie is a physical reflection of the information provided in the recipe for its making. The genetic recipe for life, written in the form of DNA, is contained in all organisms on Earth and it is passed down through the generations. Parents pass the material to their children, children to grandchildren, etc. The material includes such information as, for example, the colour of eyes or hair, shape of lips, height or intellectual aptitude. DNA is in the form of two strands on which, like on magnetic tape, a recipe for making all the proteins is written. Each strand is built from 4 types of building blocks/letters of the genetic code called nucleotides: adenosine, cytosine, guanine, and thymine. DNA strands are intertwined to form a specific spiral called a helix. The blocks suspended on both threads must be tightly matched to each other, forming adenosine – thymine, guanine-cytosine pairs. The order in which these nucleotide pairs are arranged is called the DNA sequence. The formation of a protein from the information stored in DNA is called ‘expression’. The information stored in DNA is extracted via the transcription process and finally translation results in the synthesis of the protein.

Fig. 1. DNA structure and scheme of DNA transcription into mRNA followed by mRNA translation into a polypeptide.
During transcription, information written on DNA is transcribed into RNA (so-called mRNA). RNA (ribonucleic acid), unlike DNA, is single-stranded and uses uracil instead of thymine in its genetic set of letters. During transcription, the DNA helix is temporarily unravelled. One of the strands is the template, i.e. the fundament based on which the RNA strand is built. The nucleotides of mRNA form complementary connections with the nucleotides on the DNA strand. And so, if we have the sequence adenine-guanine-cytosine-thymine-guanine on the DNA strand, the complementary RNA strand will have the sequence uracil-cytosine-guanine-adenine-cytosine. After the entire sequence has been transcribed from DNA to RNA, the helix is re-entangled back to its original form and the RNA is involved in the subsequent stages of protein production. The brick/letter sequence in RNA is a recipe for the sequence of amino acids in a protein by a process termed translation.
In the course of protein synthesis, RNA nucleotides are read in triples, and the three RNA bases encode one amino acid in the protein. These three RNA bases are called a codon, and we can imagine them as a single word in a recipe. The famous ‘genetic code’ is the rule on how to translate the sequence of nucleotides (and codon words made of them) into the sequence of amino acids which make a protein. We have 61 possible codons and any three nucleotides can code for only one amino acid (interestingly organisms use only 21 amino acids so we have more combinations at our disposal that are needed). After the translation is completed, the resulting peptide (i.e. a chain of amino acids) can be modified, another peptide can be added to it, and it can also be transported to a target location in the cell, where it can perform the functions assigned to it. In a word, it can be a building block of a cell, become a signal molecule (a hormone like insulin) or assume the role of a catalyst, that is, take the form of an enzyme [2].
Directed evolution of enzymes
The verb ‘To evolve’ is derived from the Latin as ‘evolvere’ means ‘to develop’, ‘to reveal so far undiscovered possibilities’ [8]. From the very beginning of life on Earth, enzymes have undergone a process of constant change that allows organisms to adapt to a constantly changing environment. The source of these changes is mutations that are caused by random errors that occur when DNA is copied during cell division. The selection factor is the ability of the offspring organism to survive – if it increases, a given mutation has a chance to propagate (spread) in the population. The directed evolution of enzymes is nothing more than the transfer of processes occurring in nature to the laboratory. Of course, we need to accelerate the process of change – nobody wants to wait a lifetime to see if his research worked [11]. The first reports on the successful application of this method date to the last decades of the 20th century. In 1984, Manfred Eigen published a theoretical work in which he described a possible work scheme aimed at constructing an “evolutionary machine” for optimizing already existing enzymes [6].
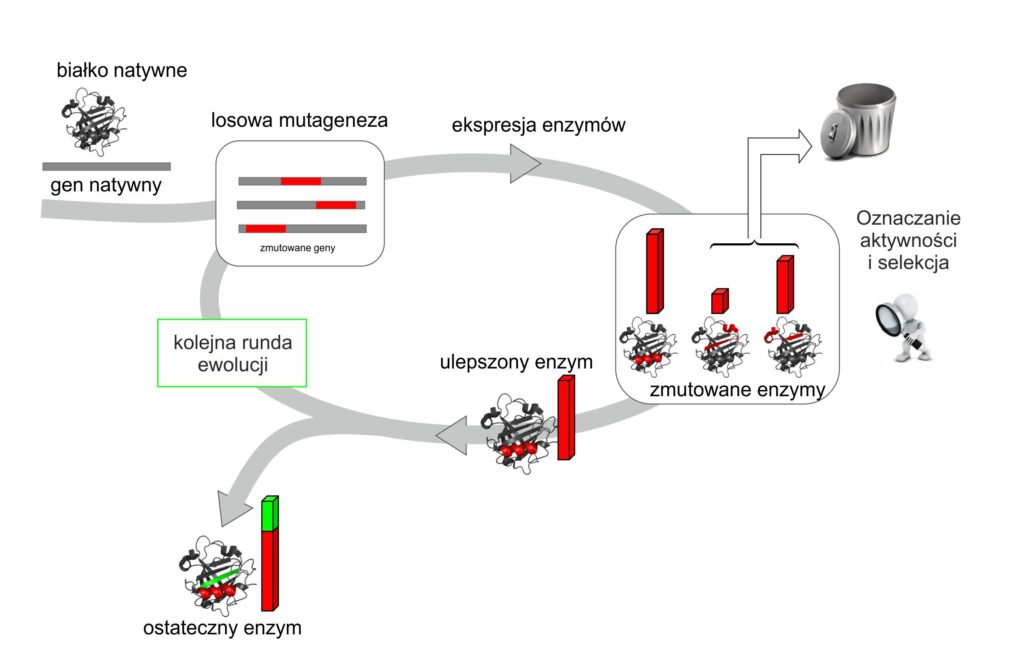
Fig. 2. Diagram showing the principle of directed enzyme evolution
The strategy of directed evolution requires the isolation of the gene encoding a given enzyme, and then copying it multiple times while introducing some variation at the level of the genetic code (i.e. provoking a mutation) (Fig. 2). Such a mutation-prone method produces many alternate copies of the original gene. The changes introduced in the gene result in a modification of the amino acid sequence of the enzyme protein of interest, which in turn leads to changes in its structure and function. This allows the creation of a library of sequences encoding different variants of the same enzyme. Coming back to the kitchen example from at the beginning of the article, some modifications are made to the dough recipe, e.g. instead of 100 grams of wheat flour, we have 130 grams, instead of 50 ml of milk, we add 70 ml, or a little cream, etc. The modification will have an impact on the structure of the biscuit. It will be firmer by increasing the amount of flour, or moister by adding more milk, or more aromatic due to cream, after all, the fat contained in cream is the flavour carrier. It should be emphasized, however, that the variability of protein sequences in the case of directed evolution is random, as opposed to rational and directed redesign, where rationally selected amino acids that make up the protein are changed. This means that in the case of a rational redesign, we know that we want to get the cake moister, so we increase the amount of milk, while in the case of targeted evolution, we create a set of recipes in which we randomly change the quantity of ingredients that make up the recipe and check how the new composition affects the appearance, structure, and taste of cookies. In the next steps, randomly changed DNA sequences are introduced into cells, either bacterial or eukaryotic. The cells are then cultured to check that e.g. the new enzyme variant has not caused destructive changes leading e.g. to a lack of growth of such a cell. Surviving cells are then tested for the expression of altered DNA sequences to check whether the introduced mutations did not, for example, block the synthesis of a new form of the enzyme. In the final stage, we obtain a library of cells that express different variants, the so-called mutant enzymes. We call a set of cells a library because each of them contains a slightly different recipe for our enzyme. Then a selection process is performed, leading to the selection of cells producing the enzyme with improved properties (e.g. by testing the enzyme’s resistance to high temperature or the ability to catalyze reactions with a new substrate). The identification of those gene variants that code for “improved” enzymes allows their further use. Usually, successive rounds of random mutagenesis and selection are carried out, which result in the accumulation of desired traits in one variant [1].
About 10 years after the theoretical article by Eigen was published in 1993, the first experimental work by Frances H. Arnold appeared. In this paper, the author described a modified variant of the subtilisin E enzyme obtained with directed evolution. Natural subtilisin E is an enzyme that catalyzes the reactions of protein hydrolysis only in a highly alkaline environment. In other words, this enzyme catalyzes the cleavage of the amino acid peptide chain in other proteins present in the cell. It should also be added that natural subtilisin E does not show catalytic activity in the presence of organic solvents, which makes its use in organic synthesis much more difficult. After several cycles of directed evolution, Frances Arnold obtained a new type of enzyme, which in the presence of 60% dimethylformamide (an organic solvent loved by organic chemists) shows 256 times higher activity compared to its naturally occurring form in the environment [5, 9]. Frances Arnold developed a complete directed evolution scheme including the steps of identifying the enzyme to be subjected to this process, techniques for introducing random mutations into the enzyme sequence, obtaining libraries of different variants of the enzyme, choosing the selection conditions and finally selecting the most preferred forms of the enzyme.
Laboratory evolution in the service of industry
Since then, scientists are more and more willing to use the methods of directed evolution, which significantly widens the range of possible applications of known enzymes as effective biocatalysts. For years this method has been applied in laboratories all over the world to obtain new efficient and environmentally friendly alternatives to the catalysts used so far. Enzyme variants resulting from directed evolution have found their application not only in many industries (e.g. food, textile, chemical or biomedical industries), but also in the household (e.g. enzymatic washing powders).
A good example of such enzymes are cellulases, which have been used both in the textile industry and in the household. They are used, among others, in the final stages of denim production, for softening cotton, or in detergents as colour-preserving ingredients. Cellulases used as a component of detergents should be compatible with other components of the washing liquid or powder, and be thermally stable stability and active at alkaline conditions present in our washing machines. Sangren and colleagues improved the temperature stability of one of the cellulases by 7.7 ° C, which means that using a detergent containing this enzyme it is possible to use it at a higher soaking or washing temperature [10].
One of the more modern challenges of science is the sustainable and environmentally friendly production of biofuels that can be an alternative to fossil fuels. The aim is to produce ready-made fuel components or building particles from which the petrochemical industry can synthesize fuels from renewable raw materials. One potential candidate for this role is isobutanol, which can either be added to gasoline or converted into short-chain alkanes. Isobutanol can be produced by biosynthesis from sugars in recombinant (i.e., genetically modified) E. coli cells. However, two enzymes of the pathway involved in its synthesis require the presence of a specific cofactor, the so-called NADPH (dinucleotide phosphate nicotinamide adenine). Unfortunately, glycolysis, which is a natural process accompanying bacterial growth, produces large amounts of another cofactor, NADH (dinucleotide nicotinamide adenine). Arnold and colleagues used directed evolution to change the dependence of both enzymes in the biosynthetic pathway on NADPH on the dependence on NADH, which enabled the production of isobutanol in bacterial cells [3].
Yet another example of the application of directed evolution at the intersection of biotechnology and biomedicine is the development of a protein proximity labelling strategy. Proximity labelling is one of the techniques for proteomic analysis of proteins. Proteomics is a branch of science that deals, in general, with everything related to proteins – that is structure of proteins, their functions, discovering the relationships that occur between proteins or between proteins and other elements in the cell and the living organism. The contactless labelling technique provides insight into the complex processes taking place in the cell. But how does it actually work? The first step is to genetically modify the genes of the protein under study (protein A) in a living organism. The modification produces the so-called fusion protein, a special enzyme (E) attached to the tested protein A. This enzyme E catalyzes the reaction of labelling other proteins. As a result of the expression of the resulting fusion protein (AE) in the natural place where the protein A occurs in the cell. The labelling takes place very close (on a nanometric scale) to the AE fusion protein is placed. In the next step, the proteins marked by the E enzyme can be easily isolated from the cell and identified. Thanks to this technique, we can find out which proteins are produced in cell organelles. We are able also to track down with which proteins the AE fusion protein has interacted. This information can be used to better design drugs or detection of metabolic diseases, in which enzymes occurring in the human body are of great importance.
This technique uses two enzymes capable of labelling proteins with biotin, ascorbate peroxidase isolated from soybeans (APEX2) and biotin ligase isolated from E. coli (BiolD). APEX2 is an enzyme that allows for very fast labelling of nearby proteins, and thus the identification of dynamic interactions taking place in living cells. Unfortunately, its use requires the use of hydrogen peroxide (H2O2). Firstly, this reagent is toxic and, secondly, it is difficult to introduce H2O2 into the cells without damaging them.
In turn, BiolD is an attractive enzyme due to the simplicity of labelling and the non-toxic conditions of this process. Unfortunately, unlike APEX2, labelling using BiolD requires much more time, i.e. 18 to 24 hours, which makes it completely impossible to observe dynamic processes taking place in cells. Brannon and colleagues using directed evolution developed two new variants of biotin ligase, TurbolD and miniTurbo, which allowed to shorten the labelling process from many hours to about 10 minutes. It also became possible to use this type of labelling in yeast cells (i.e. in eukaryotic cells similar to ours), which was impossible before [4].
Targeted evolution has also become a business idea. There are several companies on the world markets that specialize in improving enzymes already used in industry and creating new proteins that can be used commercially. Such companies include, for example, ATUM Company, GeneArt Directed Evolution operating under the name ThermoFischer Scientific, or Creative Enzymes [12 – 14].
Summary
It has been almost 35 years since the directed enzyme evolution method was discovered. Although this method allows the creation of enzymes that operate even faster or under different conditions, without the need to fully understand the relationship between enzyme structure and function, there is still plenty of room for further discoveries. When journalists asked professor Arnold what advice she has for people who strive to achieve great things, she replied that the most important thing is to like what they are working on, day by day and to take risks, because no one who has achieved success has followed the “recipe”.
.
Bibliography:
- Arnold FH. (2018). Enzymes by Evolution: Bringing New Chemistry to Life. Molecular Frontiers Journal 2: 9 – 18.
- Barciszewski, J; Markiewicz, WT. (2002). “Kwasy nukleinowe. Kod genetyczny.”
- Bastian, S; Liu, X; Meyerowitz, JT; Snow, CD; Chen, MMY; Arnold, FH. (2011). Engineered ketolacid reductoisomerase and alcoholdehydrogenase enable anaerobic 2-methylpropan-1-ol production a theoretical yield in Escherichia coli. Metabolic Engineering 13: 345 – 352.
- Branon, TC; Bosch, JA; Sanchez, AD; Udeshi, ND; Svinkina, T; Carr, SA; Feldman, JL; Perrimon, N; Ting, AY. (2018). Efficient proximity labeling in living cells and organisms with TurboID. Nature Biotechnology 36: 880 – 887.
- Chen, K; Arnold, FH. (1993). Tuning the activity of an enzyme for unusual environments: sequential random mutagenesis of subtilisin E for catalysis in dimethylformamide. Proceedings of the National Academy of Sciences of the United States of America 90: 5618 – 5622.
- Eigen, M; Gardiner, W. (1984). Evolutionary molecular engineering based on RNA replication. Pure and Applied Chemistry 56: 967 – 978.
- Hult, K; Berglund, P. (2003). Engineered enzymes for improved organic synthesis. Current Opinion in Biotechnology 14: 395 – 400.
- Kłys, A. Słownik łacińsko – polski, polsko – łaciński. Wydanie 1. (2012). Wydawnictwo Level Trading, Czerica.
- Kuchner, O; Arnold, FH. (1997). Directed evolution of enzyme catalysts. Trends in Biotechnology 15: 523 – 530.
- Sandgren, M; Gualfetti, PJ; Shaw, A; Gross, LS; Saldajeno, M; Day, AG; Jones, TA; Mitchinson, C. (2003). Comparison of family 12 glycoside hydrolases and recruited substitutions important for thermal stability. Protein Science 12: 848 – 860.
- Scientifc Background on the Nobel Prize in Chemistry 2018, Stockholm, doi: https://www.nobelprize.org/uploads/2018/10/advanced-chemistryprize-2018.pdf.
- www.atum.bio/company
- www.creative-enzymes.com/
- www.thermofisher.com/pl/en/home/life-science/cloning/gene-synthesis/directed-evolution.html